Processing large CSV files
Just when I thought I would be good with importing a large CSV file into Postgresl using csvkit, I realized that nothing was going as planned: It was taking lot of memory and it was going on far too long when I would like to avoid this kind of inconvenience by transferring a CSV file to a database.
The CSV files I am interested in come from the National Bureau of Economic Research (I introduced them in an earlier post), and they are quite big. For instance, the dataset about vital statistics from 2016 is composed of almost 4 millions of records:
natl2016 ll
total 11236760
-rw-rw----@ 1 chl staff 2,2G 4 oct 2017 natl2016.csv
-rw-r--r--@ 1 chl staff 27K 4 jan 2018 natl2016.dct
-rw-r--r--@ 1 chl staff 1,2K 4 jan 2018 natl2016.do
-rw-r--r--@ 1 chl staff 966M 4 oct 2017 natl2016.dta
-rw-r--r--@ 1 chl staff 1,5M 4 jan 2018 natl2016.pdf
-rw-rw----@ 1 chl staff 2,2G 19 sep 18:50 natl2017.csv
natl2016 wc -l natl2016.csv
3956113 natl2016.csv
natl2016
As I had no luck with csvsql on the whole file (with bare default options, that is without disabling type inference or constraints checking), I thought it could be a good idea to split up the file in chunks of more resaonnable size, say 500,000 records each. Here it comes slowly: We need some efficient text processing utilities. Here is a little attempt at using sed to extract the first three rows. Note that we keep the header since it will be used to create variable names in the table when the file is processed by csvsql:
natl2016 sed -n 1,4p natl2016.csv > out.csv
natl2016 csvsql --db postgresql://localhost/surveys --table natl2016 --insert out.csv
So far so good, it works like a charm.
We could use a little loop to ask sed to do that 5 to 10 times, but split is exactly what we are looking for. It is also more flexible as we can decide to split a file into chunks of given length (-l) or size (-b). In this case, it is not possible to split in chunks of fixed size since this could result in incomplete lines. So we will be using a fixed length for each chunk (500,000 rows). Here it is in action:1 (the last line is just to ensure that we haven’t lost anything during the process)
natl2016 split -l500000 natl2016.csv part_
natl2016 mkdir out && cd $_
natl2016 mv ../part_* .
natl2016 find . -name 'part_*' | xargs wc -l
The same result could be achieved using Python and a slicing pattern from the itertools package (this is certainly slower but it will keep you happy with the memory pressure). The read_csv function from Pandas also offers an option to handle large files with chunking. However, no tool will beat the above command, IMHO.
To import the data files into the Postgresql database, we have two “standard” options: csvsql or a BULK INSERT statement. Another option would be the recently released d6tstack Python package, which has the advantage to work with Excel files as well while offering more backends (Pandas data frames or Parquet files). Some benchmarks are available on Github. Obviously, we would prefer not to have to recreate the schema, even if it is not that difficult given that the NBES also provides a Stata dictionnary for those data. I yet have to write a “dct2sql” translator! Anyway, csvsql which seemed to be a good candidate cannot be uses in “append” mode, so we are done with Python. Moreover, after experimenting a little, with or without type inference, I think it is not a practical solution since it is still very slow. Likewise, my initial tests with d6tstcack were not as promising as I expected.
There are, however, faster ways to perform bulk insertions in Postgresql, including pg_bulkload and most importantly the built-in COPY FROM statement. But let’s that not prevent us from continuing with GNU tools. In what follows, we will take a random sample of the data file to ensure better type inference than the first few lines. We also need to append the CSV header to get variable name, and remove it from the first file generated by split (gsed is GNU sed for Mac users).2 It is also important to ensure that empty strings would not raise errors on import so we take care of that in the schema file directly (otherwise we would have to post-process each file using complex regex patterns). Note that the table has a strange name (stdin); this comes from our piping operations, which prevent us to give the table a proper name. Let’s rename it:
natl2016 head -1 natl2016.csv > header
natl2016 shuf -n 10000 natl2016.csv | cat header - | csvsql > schema
natl2016 gsed -i '1d' out/part_aa
natl2016 gsed -i 's/ NOT NULL//g' schema
natl2016 gsed -i 's/stdin/natl2016/' schema
natl2016 cat schema | psql -d postgresql://localhost/surveys
Finally, we can import the data using psql directly. A single query runs in just under 30 seconds on my Macbook (1.3 GHz Intel Core m7), so we could safely loop over all files and populate the database:
for f in out/part_*
do
echo "Loading $f"
psql -d postgresql://localhost/surveys -c "\copy natl2016 FROM '$f' (FORMAT csv)"
done
That’s it. Here are the results from Postico:3
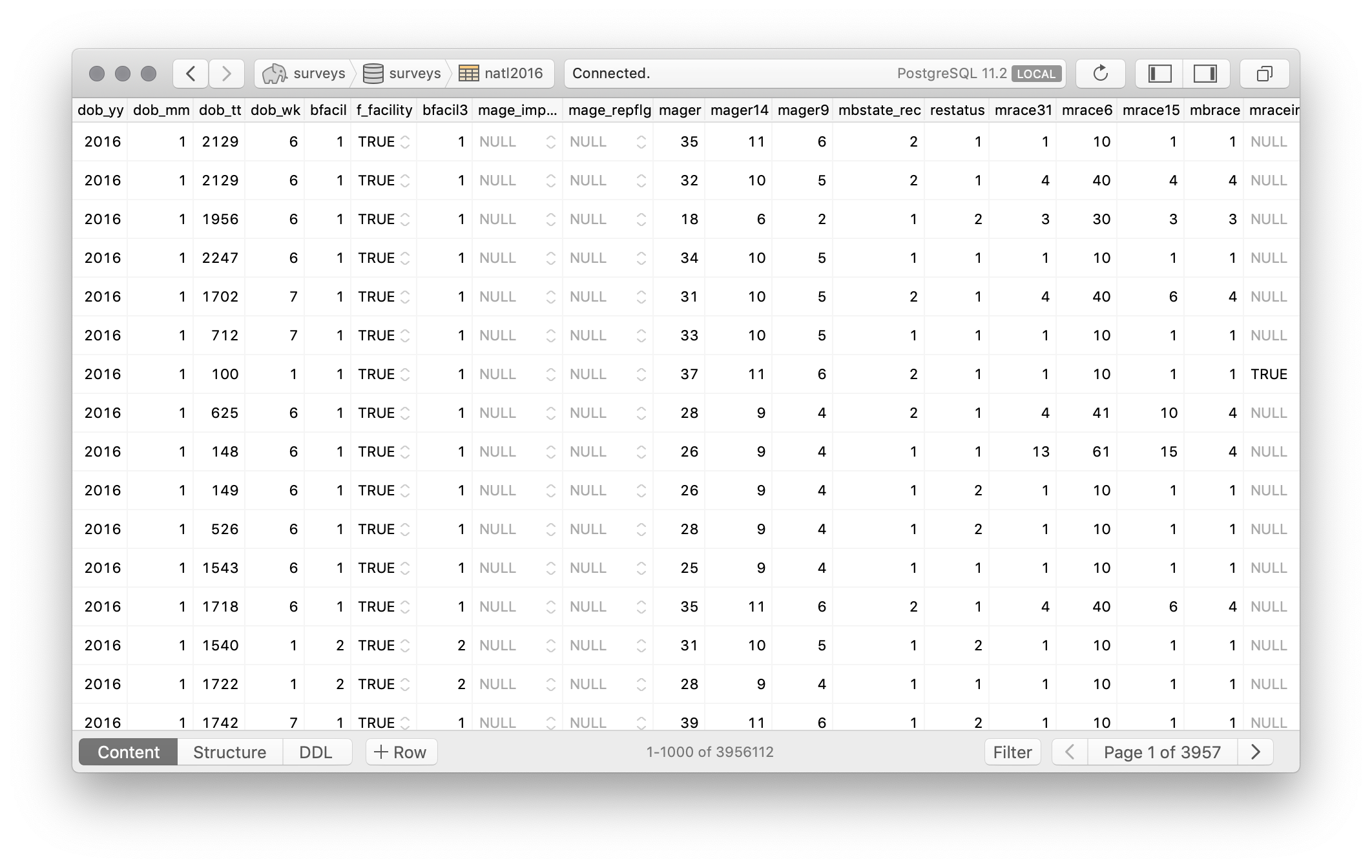
Now, I believe it should be possible to input the whole CSV file but I haven’t tried.
surveys=# select sex, count(*) from natl2016 group by sex;
sex | count
-----|---------
F | 1932663
M | 2023449
(2 rows)
Notice that this table does not have a primary key (it might be a good idea to add one at some point since it may significantly improve performance on grouped-by queries) and the dob_yy column can be safely removed since it is just the year of the survey. In a next post, we shall take a closer look at built-in extensions and window functions.
-
(Initially, I though about piping like this:
cat schema | sed "1 s/^/'/; $ s/$/'/" | psql -d postgresql://localhost/surveys -c. This adds surrounding single quotes to the output ofcatbefore piping the result topsql. This doesn’t work becausepsqlcan work with pipes but we don’t need the-coption in this case. Other solutions to insert SQL statements do exist, like named pipes.) ↩︎ -
Viewing the data takes 30s. ↩︎