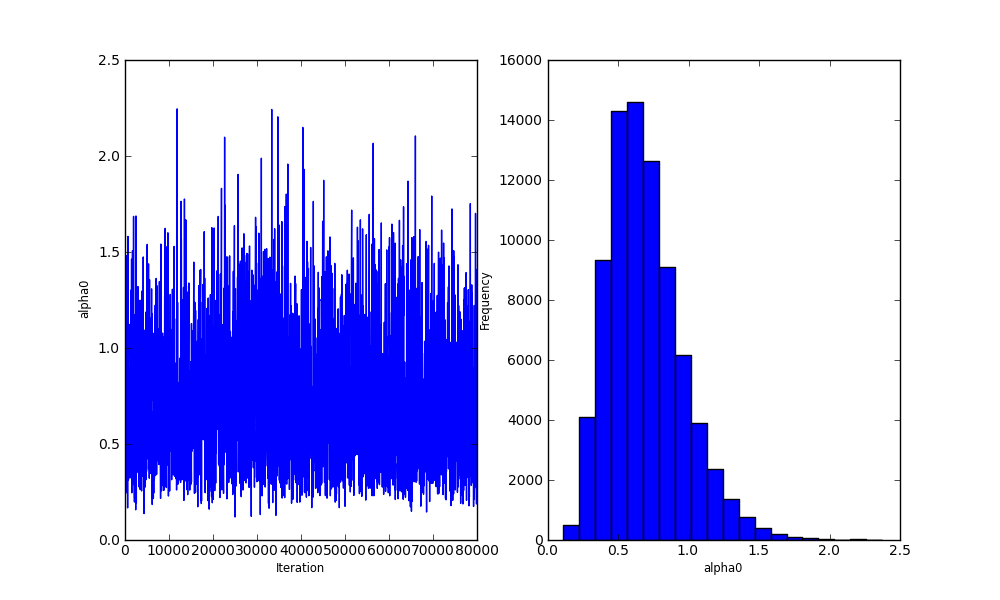
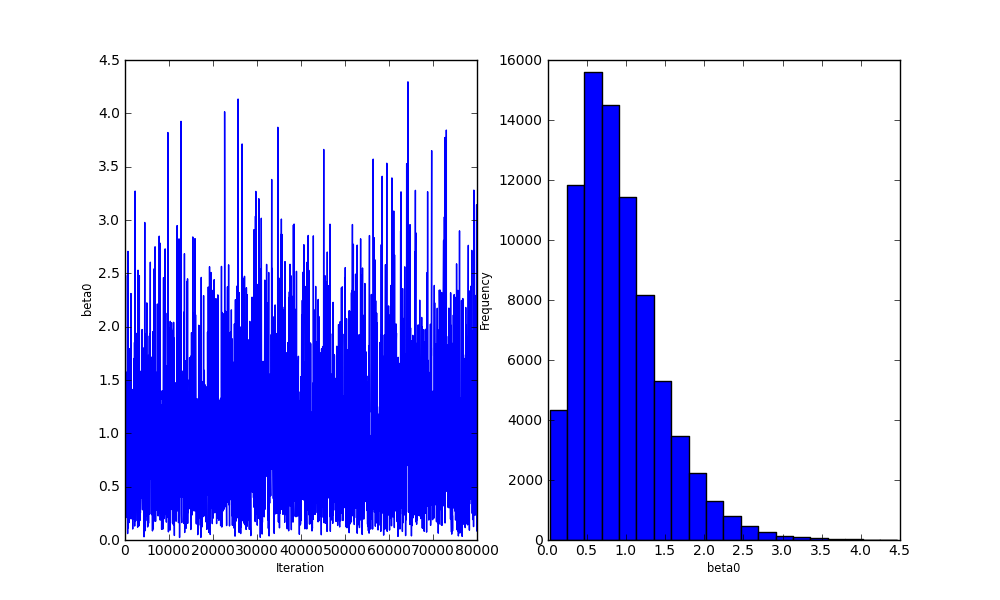
Bayesian analysis with Python
Aside from my previous post on carrying out Bayesian analysis with R, here is an illustration on a Hierarchical Poisson failure rates from Clark and Gelfand,1 using Python and the PyMC package.
The Python code is as simple as the R code, although it is obviously more object-oriented. The main part are highlighted below. These are in fact where we specify the prior distributions
alpha0 = Exponential('alpha0', 1.0, value=1.)
beta0 = Gamma('beta0', alpha=0.1, beta=1.0, value=1.)
theta = Gamma('theta', alpha=alpha0, beta=beta0, value=ones(k))
Running the model amounts to:
var_list = [alpha0, beta0, theta, y]
M = MCMC(var_list)
M.use_step_method(AdaptiveMetropolis, [alpha0, beta0])
M.isample(100000,burn=20000,thin=1,verbose=2)
Matplot.plot(M)
Sidenote
As of June, the 27th, there are problem with the gp package (gp_submodel.py isn’t in the source distribution, nor does it comes from the Universal installer through pipy). This is discussed on one Google group thread. I happened to setup a working version of PyMC by downloading the source from the Github repository and compiling from scratch, with the following commands:
$ export LDFLAGS="-Wall -lgfortran -undefined dynamic_lookup -bundle -arch x86_64"
$ python setup.py config_fc --fcompiler gnu95 build
$ sudo python setup.py install
Clark, J.S. and Gelfand, A. (2006). Hierarchical modelling for the environmental sciences: statistical methods. Oxford university Press. ↩︎